Out-of-distribution prediction with disentangled representations for single-cell RNA sequencing data
Abstract
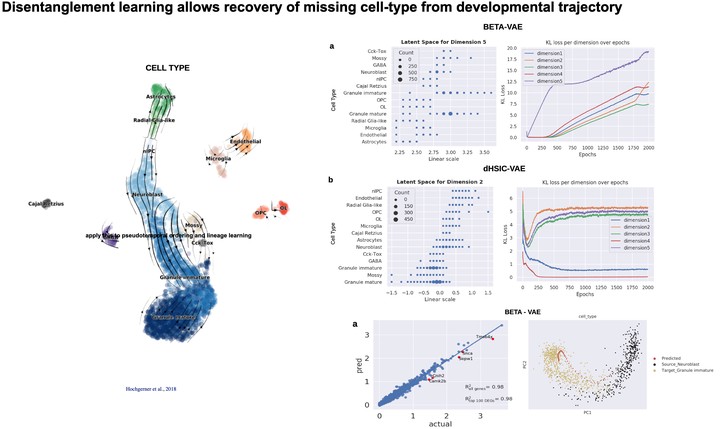
Learning robust representations can help uncover underlying biological variation in scRNA-seq data. Disentangled representation learning is one approach to obtain such informative as well as interpretable representations. Here, we learn disentangled representations of scRNA-seq data using β variational autoencoder (β-VAE) and apply the model for out-of-distribution (OOD) prediction. We demonstrate accurate gene expression predictions for cell-types absent from training in a perturbation and a developmental dataset. We further show that β-VAE outperforms a state-ofthe-art disentanglement method for scRNA-seq in OOD prediction while achieving better disentanglement performance.
Type
Publication
ICML workshop in Computational Biology, spotlight talk, poster